
Plot two-dimensional data modelled by an MVN mixture
mclust2Dplot.RdPlot two-dimensional data given parameters of an MVN mixture model for the data.
Usage
mclust2Dplot(data, parameters = NULL, z = NULL,
classification = NULL, truth = NULL, uncertainty = NULL,
what = c("classification", "uncertainty", "error"),
addEllipses = TRUE, fillEllipses = mclust.options("fillEllipses"),
symbols = NULL, colors = NULL,
xlim = NULL, ylim = NULL, xlab = NULL, ylab = NULL,
scale = FALSE, cex = 1, PCH = ".",
main = FALSE, swapAxes = FALSE, ...)Arguments
- data
A numeric matrix or data frame of observations. Categorical variables are not allowed. If a matrix or data frame, rows correspond to observations and columns correspond to variables. In this case the data are two dimensional, so there are two columns.
- parameters
A named list giving the parameters of an MCLUST model, used to produce superimposing ellipses on the plot. The relevant components are as follows:
proMixing proportions for the components of the mixture. There should one more mixing proportion than the number of Gaussian components if the mixture model includes a Poisson noise term.
meanThe mean for each component. If there is more than one component, this is a matrix whose kth column is the mean of the kth component of the mixture model.
varianceA list of variance parameters for the model. The components of this list depend on the model specification. See the help file for
mclustVariancefor details.
- z
A matrix in which the
[i,k]th entry gives the probability of observation i belonging to the kth class. Used to computeclassificationanduncertaintyif those arguments aren't available.- classification
A numeric or character vector representing a classification of observations (rows) of
data. If present argumentzwill be ignored.- truth
A numeric or character vector giving a known classification of each data point. If
classificationorzis also present, this is used for displaying classification errors.- uncertainty
A numeric vector of values in (0,1) giving the uncertainty of each data point. If present argument
zwill be ignored.- what
Choose from one of the following three options:
"classification"(default),"error","uncertainty".- addEllipses
A logical indicating whether or not to add ellipses with axes corresponding to the within-cluster covariances.
- fillEllipses
A logical specifying whether or not to fill ellipses with transparent colors when
addEllipses = TRUE.- symbols
Either an integer or character vector assigning a plotting symbol to each unique class in
classification. Elements incolorscorrespond to classes in order of appearance in the sequence of observations (the order used by the functionunique). The default is given bymclust.options("classPlotSymbols").- colors
Either an integer or character vector assigning a color to each unique class in
classification. Elements incolorscorrespond to classes in order of appearance in the sequence of observations (the order used by the functionunique). The default is given ismclust.options("classPlotColors").- xlim, ylim
Optional argument specifying bounds for the ordinate, abscissa of the plot. This may be useful for when comparing plots.
- xlab, ylab
Optional argument specifying labels for the x-axis and y-axis.
- scale
A logical variable indicating whether or not the two chosen dimensions should be plotted on the same scale, and thus preserve the shape of the distribution. Default:
scale=FALSE- cex
An argument specifying the size of the plotting symbols. The default value is 1.
- PCH
An argument specifying the symbol to be used when a classificatiion has not been specified for the data. The default value is a small dot ".".
- main
A logical variable or
NULLindicating whether or not to add a title to the plot identifying the dimensions used.- swapAxes
A logical variable indicating whether or not the axes should be swapped for the plot.
- ...
Other graphics parameters.
Value
A plot showing the data, together with the location of the mixture components, classification, uncertainty, and/or classification errors.
Examples
# \donttest{
faithfulModel <- Mclust(faithful)
mclust2Dplot(faithful, parameters=faithfulModel$parameters,
z=faithfulModel$z, what = "classification", main = TRUE)
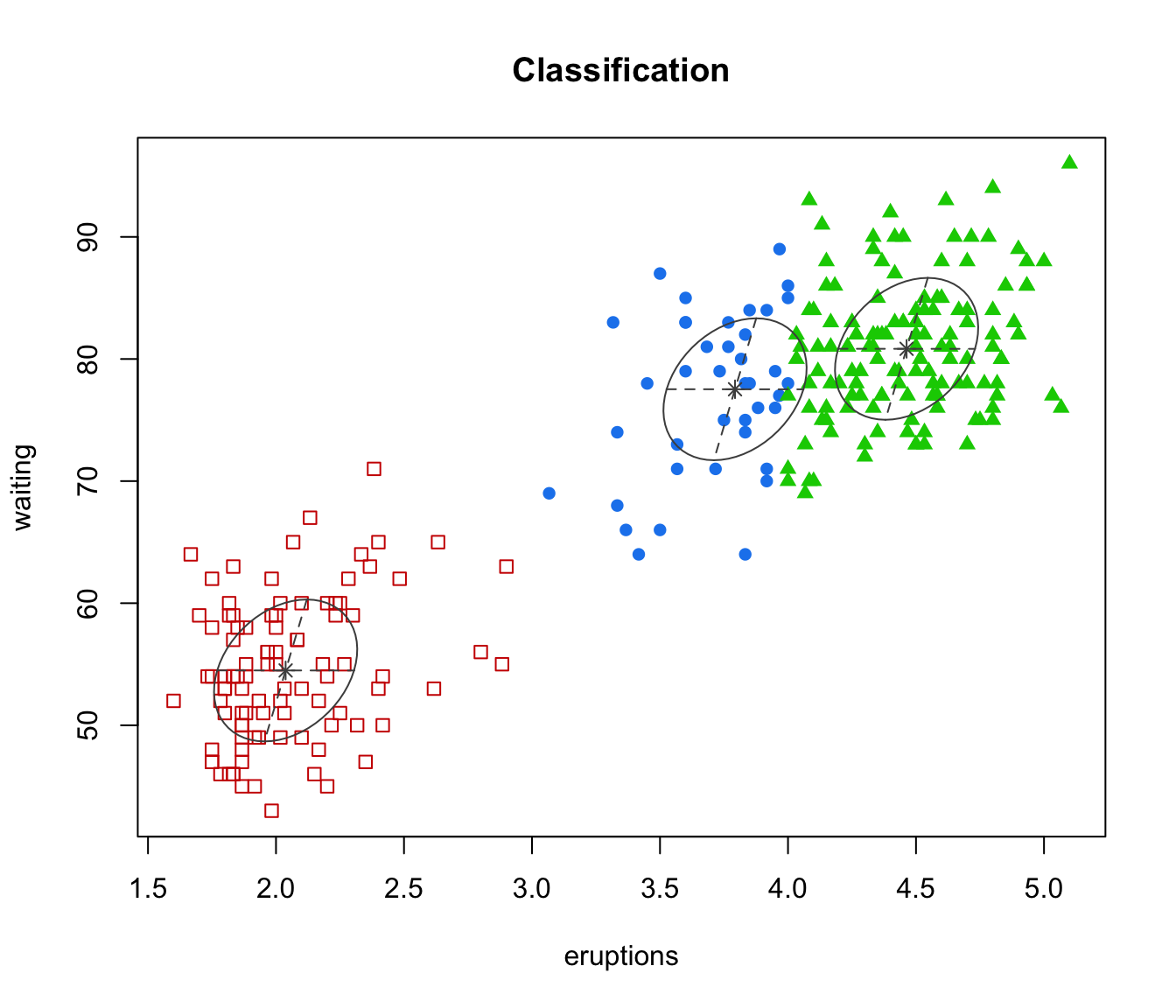 mclust2Dplot(faithful, parameters=faithfulModel$parameters,
z=faithfulModel$z, what = "uncertainty", main = TRUE)
mclust2Dplot(faithful, parameters=faithfulModel$parameters,
z=faithfulModel$z, what = "uncertainty", main = TRUE)
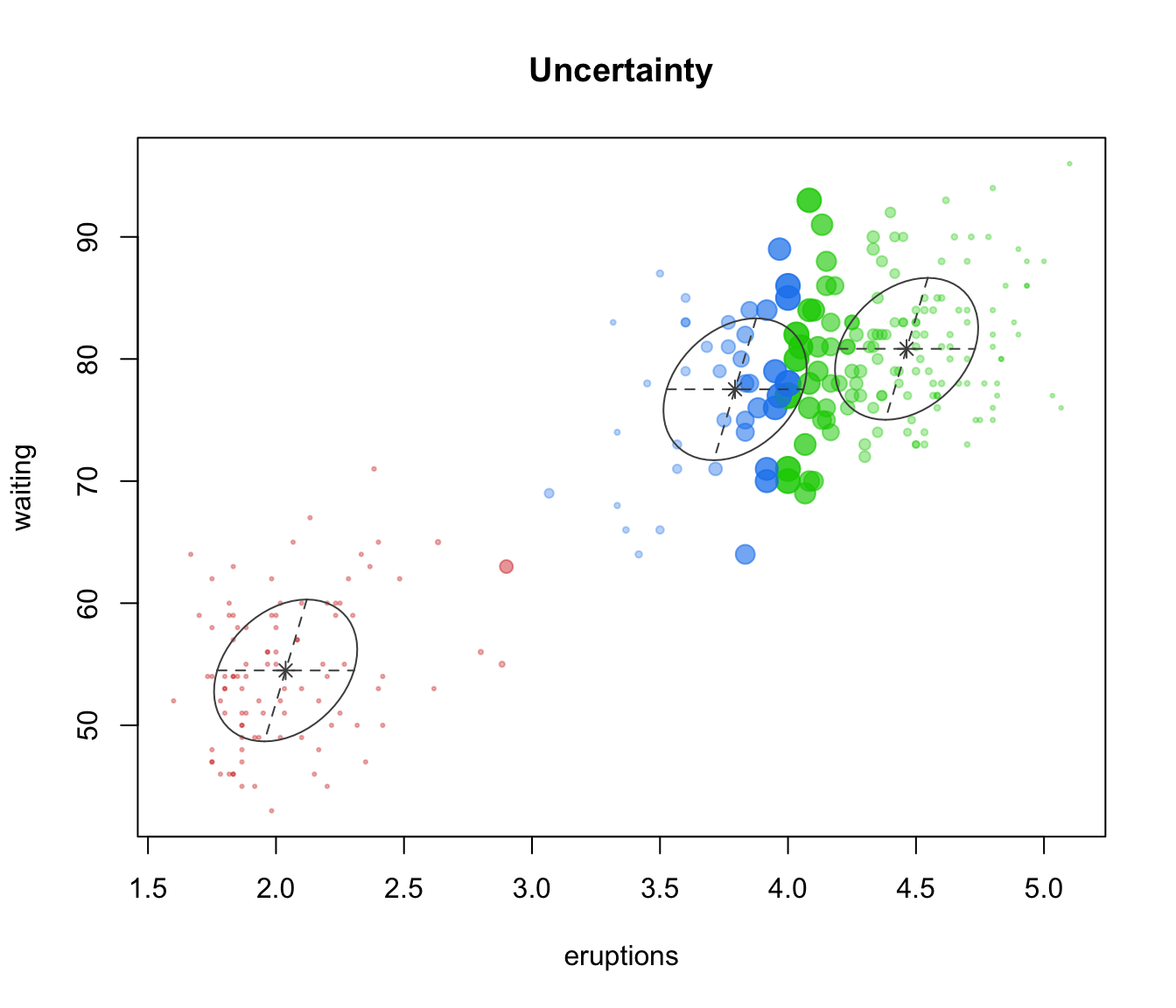 # }
# }